Weijian Yang, Columbia University
THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
A new imaging tool creates a hologram of neurons firing across distant areas of the mouse brain. The tool, described in two papers published 20 January in Neuron, could reveal how brain circuits are altered in mouse models of autism1,2.
Other instruments allow researchers to watch neurons fire within a thin layer of brain tissue by tracking the flow of calcium into the cells. To obtain a more complete view of this activity, researchers must zoom and refocus the microscope on progressively deeper tissues, then merge pictures from each depth, or layer, to create a three-dimensional (3D) image.
The new tool consolidates this process by scanning several layers at once and automatically generating a 3D rendering. Not only does the tool save time, but its ability to scan many layers at a single point in time allows researchers to observe synchronized firing in distant regions of the mouse brain.
To create the tool, the researchers equipped a fluorescent microscope with a device that splits a single laser beam into smaller ‘beamlets.’ These beamlets scan different depths of brain tissue and detect the light emitted by fluorescent indicators when calcium flows into firing neurons.
Data from the modified microscope feed into an algorithm that builds a hologram of a section of brain tissue as deep as 700 micrometers, about the thickness of a grain of salt. The entire process unfolds in the amount of time needed to scan a single layer of brain tissue using traditional techniques.
Test drive:
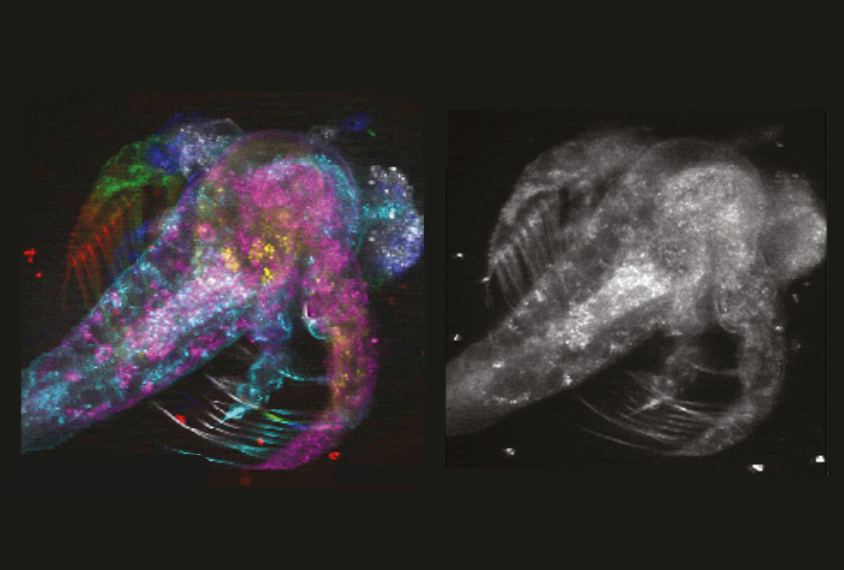
The researchers tested the device on the naturally fluorescing tissue of a living brine shrimp, gathering images from seven depths of structural tissue as the tiny animal floated in a dish. They compared the device with two existing tools: one that stitched sequential snapshots from seven layers of the shrimp’s tissue into a 3D portrait, and another that used beamlets to capture the snapshots simultaneously but required researchers to manually merge the images.
The new device generated an instant snapshot of neuronal activity in all seven layers, with image quality comparable to that of existing tools.
The researchers also used the tool to scan different depths of brain tissue in a living mouse. They immobilized the animal’s head under the microscope as the mouse walked on a circular treadmill, then directed the beamlets to specific depths based on information about the location and firing patterns of distinct neuron types. The resulting holograms revealed synchronized firing among the cells that sequential scans of each layer might have missed.
By dialing up the power of certain beamlets and dialing down those focused on neighboring layers, the researchers could zoom in on neurons communicating across different depths of the primary visual cortex.
By joining the discussion, you agree to our privacy policy.