THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
Two new resources catalog how DNA variants affect gene expression in the brain; one of the resources also focuses on chemical modifications to chromosomes.
The projects could help reveal how variants contribute to conditions such as autism.
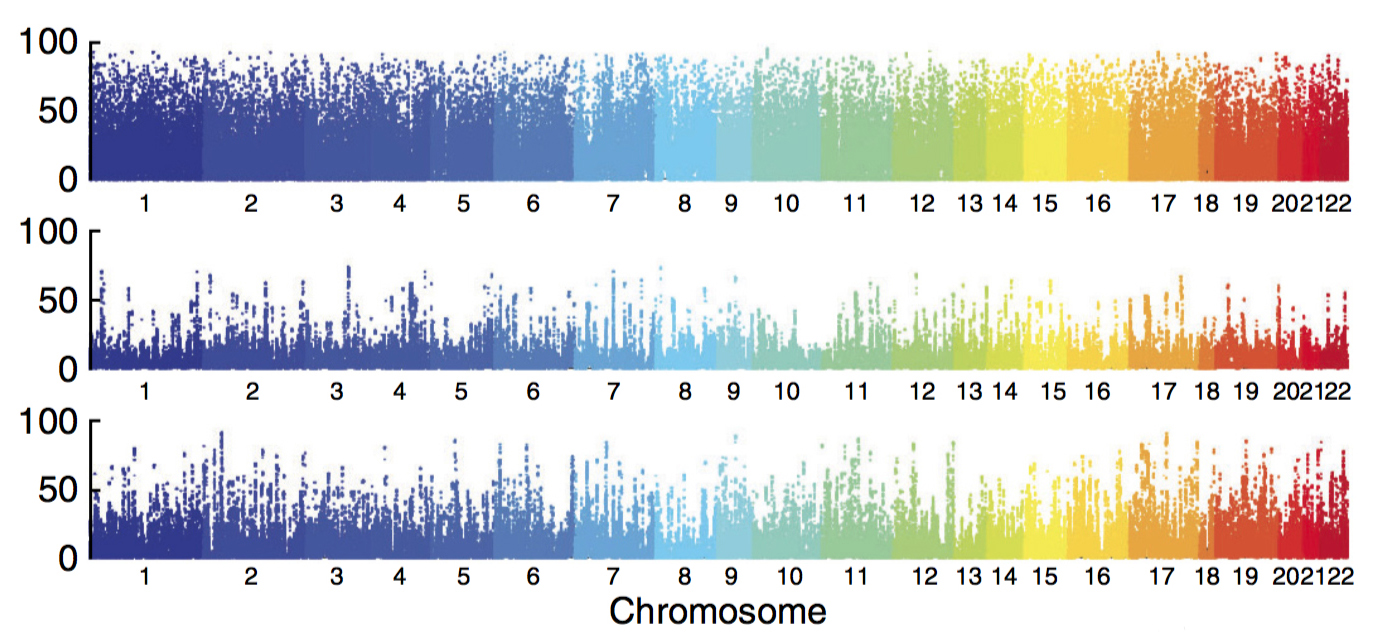
The Genotype-Tissue Expression (GTEx) project details the effects of variants on gene expression across 44 human tissue types, including the brain1. It was described last week in Nature.
The researchers analyzed DNA using postmortem blood samples from 450 people. They also extracted RNA from 7,051 tissue samples, which came from 10 brain regions, 31 other organs, blood and two cell lines derived from blood or skin samples. This provided a snapshot of the active genes in each tissue type.
The team then applied a method called quantitative trait locus (QTL) analysis to each person’s DNA and RNA. The technique links variants that land outside of genes to changes in gene expression.
They found that variants near a gene are more likely to affect its expression. And variants that control a gene’s expression in one region of the brain are more likely to control it elsewhere in the brain than in other parts of the body.
Some of the variants the study flagged were also identified in one or more genome-wide association studies (GWAS) as being unusually common in people with certain conditions.
Researchers can access the data for free at the GTEx Portal and apply them to their own GWAS findings.
Aging resource:
The other resource is a list of genetic variants associated with either changes in gene expression or chemical modifications to chromosomes that control certain genetic regions2.
Researchers matched DNA sequences with data from postmortem tissue samples from about 500 people who participated in two longitudinal studies of aging. The tissue came from the dorsolateral prefrontal cortex, a region associated with attention, reasoning and decision-making.
The team sequenced RNA in the tissues and measured two chemical modifications that affect gene expression: the addition of methyl groups to DNA, and of acetyl groups to protein ‘spools’ called histones around which DNA winds. Both changes affect gene expression by making genes less or more accessible to the cell’s DNA-reading machinery.
The researchers used QTL analysis to uncover associations between variants and gene expression, DNA methylation or histone modification. This revealed thousands of variants likely to affect gene expression in the brain, the researchers reported 4 September in Nature Neuroscience.
The associations are listed online in a new resource called xQTL Serve.
By joining the discussion, you agree to our privacy policy.