Courtesy of OIST
THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
A software program can simulate a neuron with realistic 3-D detail in a matter of hours, researchers reported Saturday at the 2017 Society for Neuroscience annual meeting in Washington D.C.
The free software, called STEPS, can be used to mimic many aspects of the properties of neurons. For example, it can simulate the membrane proteins that moderate the flow of ions. It also takes into account the complex shape of the neuron and its dendritic spines, the small protrusions that receive electric impulses from other neurons. This can help researchers study neuronal behavior in much more detail than was previously possible.
Detailed virtual neurons can also help researchers study conditions such as autism. Mutations in several autism risk genes, including SHANK3 and FMRI, result in altered dendritic spine morphology.
“It may be useful in models that try to explain the consequences of molecular defects for neuron function,” says lead investigator Erik De Schutter, head of the Computational Neuroscience Unit at the Okinawa Institute of Science and Technology in Japan.
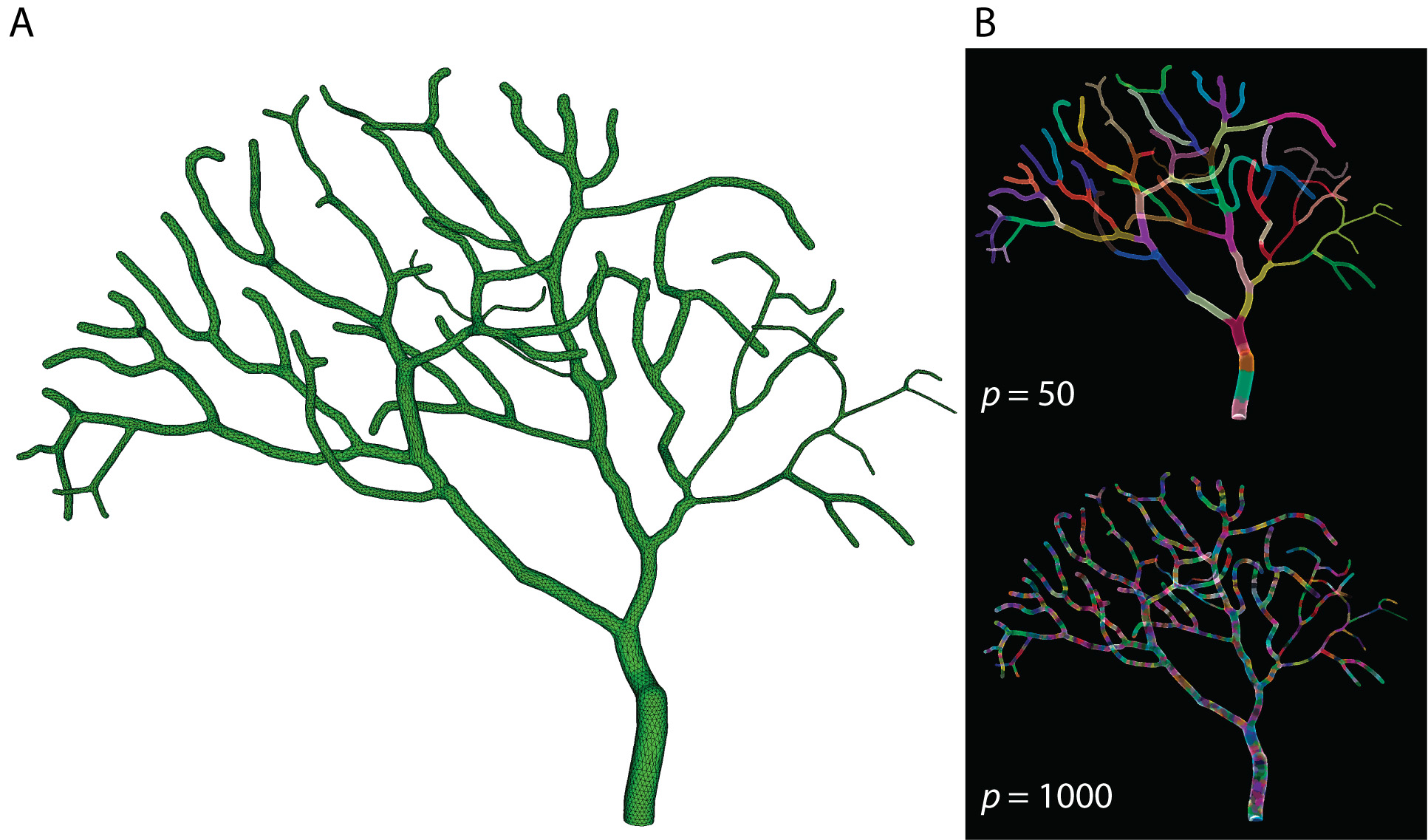
To develop the software, the researchers began with a difficult example: a Purkinje cell, a type of neuron characterized by its uniquely large number of dendrites. They were able to simulate the cell’s entire dendritic tree to sub-micron resolution.
“This is like a gold standard. If you can simulate this neuron, you can simulate any type of neuron,” says Haroon Anwar, a postdoctoral fellow at the New Jersey Institute of Technology. (Anwar was not involved in the new work but presented the model on behalf of his former colleagues.)
Computational cost:
Simulating a single neuron in full detail requires such immense computational power that it has been essentially impractical for researchers to attempt. But there are work-arounds: The conventional way to simulate a neuron has long been to assume that its dendritic spines have a smooth, cable-like structure and to use simplified mathematics for describing the behavior of ion channels.
But some functions of the neurons, such as the opening and closing of the ion channels, require computationally expensive models that simulate randomly changing systems. STEPS doesn’t shy away from them. “We simulate every single channel, as opposed to the average current being simulated in most neural models,” De Schutter says.
The team performed the computations in parallel across 2,000 computing cores, the equivalent of 1,000 computers.
The first version of STEPS was released in 2012. In this latest version, simulations that required weeks to run can be completed in a few hours.
The researchers developed the software in collaboration with the Blue Brain Project, an ongoing international project to build a computer-simulated brain, based at the École Polytechnique Fédérale de Lausanne in Switzerland.
For more reports from the 2017 Society for Neuroscience annual meeting, please click here.
By joining the discussion, you agree to our privacy policy.