THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
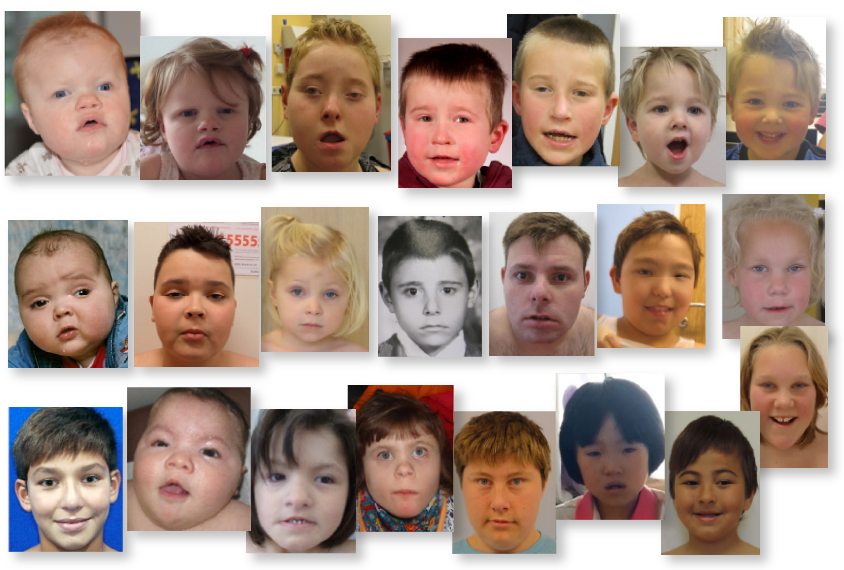
Mutations in a gene called POGZ lead to a constellation of traits, such as vision problems, hyperactivity and a small head, according to a study of 25 people with mutations in the gene1. All of the individuals have developmental delay and more than half have autism.
The findings stem from an international effort to track down and characterize as many people as possible with rare mutations in genes with strong ties to autism. Describing the diverse effects of these mutations is the first step toward identifying subtypes of autism, which might help researchers develop targeted treatments, says lead researcher Evan Eichler.
“The POGZ study highlights the power of global networking to uncover subtle but reproducible features of autism subtypes,” says Eichler, professor of genome sciences at the University of Washington in Seattle. The study appeared 3 March in the American Journal of Human Genetics.
POGZ encodes a little-understood protein that regulates the expression of other genes. It emerged as a strong autism candidate after researchers sequenced protein-coding genomic regions in thousands of people with autism. A 2014 study co-led by Eichler identified two individuals with autism who carry spontaneous, harmful mutations in POGZ.
Eichler and his colleagues then joined forces with a network of genetic-testing clinics spanning seven countries, including China and Australia. They sequenced POGZ in more than 12,000 individuals who have either intellectual disability or autism from an unknown cause. They combined their findings with those of a clinical genetics laboratory at Radboud University Nijmegen Medical Center in the Netherlands to identify a total of 25 people with harmful POGZ mutations.
The researchers found only two harmful POGZ mutations in a database of more than 45,000 people from general population.
“It’s an amazing way to be able to look at this,” says Ellen Hanson, director of the Neurodevelopmental Disorders Phenotyping Program at Boston Children’s Hospital, who was not involved in the work.
Diving deeper:
The new work comes on the heels of a smaller study published earlier this year that described five individuals with harmful POGZ mutations, all of whom have intellectual disability or developmental delay2. The individuals share subtle physical features, such as a small head — a condition called microcephaly — flat cheekbones and a flat nose.
The findings suggested that POGZ mutations underlie an intellectual disability syndrome that is characterized by these physical traits and only sometimes accompanied by autism, says Vernon Sutton, professor of molecular and human genetics at Baylor College of Medicine in Houston, Texas, who led that study.
The new work identifies additional features associated with POGZ mutations. Of the 25 individuals, 19 had their DNA sequenced because of an intellectual disability diagnosis and 6 had an autism diagnosis. The researchers had permission to contact each individual and invite them back to the clinic.
Detailed examinations revealed that all 25 have intellectual disability or developmental delay and 13 have autism; 4 others have some features of autism but do not have a diagnosis.
As in the earlier study, the scientists also found that people with POGZ mutations tend to have unusually small heads. They also identified hyperactivity, vision problems and sleep disturbances as common in the group. Two of these features — hyperactivity and vision problems — as well as small head size are more common in the group with POGZ mutations than in a database describing 2,000 people with autism.
This finding suggests that the traits are specific to a subtype of autism caused by POGZ mutations, says Holly Stessman, a postdoctoral fellow in Eichler’s laboratory. “We’re trying to pick out individuals who come to us from this broad characterization [of autism] and say, ‘We think that you have these very specific pieces of this puzzle,’” she says.
Weighty clue:
In speaking with these individuals and their families, the researchers uncovered another unusual trait: Many children with POGZ mutations have an aversion to foods with certain textures, leading to frequent vomiting and an inability to gain weight. Children who do not have the texture trouble tend to be obese, perhaps because POGZ affects metabolism, Stessman says.
The researchers identified one girl who had trouble gaining weight at first, but became obese after age 9. Another girl who is obese has a typically developing sister close to her age who is slimmer. Her diet is the same as her sister’s, suggesting that the mutation underlies her weight problem.
POGZ expression spikes during fetal development in the pituitary gland, which regulates the body’s metabolism, bolstering the theory that POGZ mutations alter metabolism.
The discovery highlights the value of talking to families, says Stessman. “It really opened our eyes,” she says. “We weren’t thinking about the pituitary at all before.”
The next step is to look at the effects of POGZ mutations in animal models. To start, the researchers deleted the equivalent gene in fruit flies. Like typical flies, the mutants jump in response to a flash of light.
Typical flies eventually stop being startled after repeated bursts of light, but the mutants never learn, continuing to jump every time — a behavior previously associated with mutations in intellectual disability genes. The findings hint that mutations in POGZ may cause intellectual disability in people.
By joining the discussion, you agree to our privacy policy.