Guillaume Auzias
THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
Combining data from different brain scanners can lead to false findings if variation between the machines is not taken into account, suggests a new study. The work helps explain why results detailing the neuroanatomy of autism often contradict one another or are not replicated1.
Merging imaging data from different labs offers a possible solution to a perennial problem: recruiting enough participants to get a statistically significant result. Yet scanners vary along dimensions such as the strength of the magnetic field they generate. What’s more, research teams may use different programs to extract anatomical information from the images.
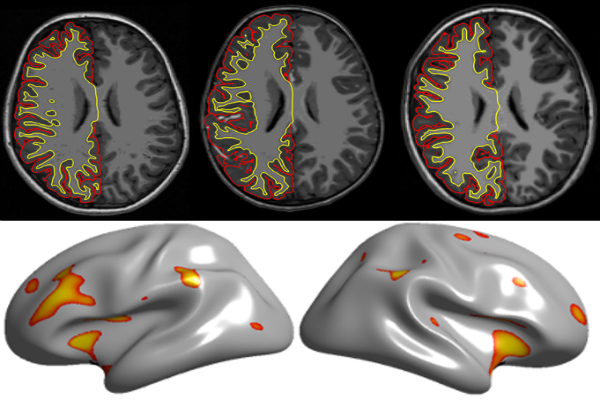
Although investigators who use magnetic resonance imaging (MRI) to examine brain structure knew about this variability, until now, no one had quantified it in studies of autism or in children.
Christine Deruelle and her colleagues found that discrepancies in the measurements from scanners at different imaging centers can introduce huge variation in the data about brain structure in autism. The results were published 22 July in the IEEE Journal of Biomedical and Health Informatics.
“For the first time, we show that these recording parameters have a major effect,” says Deruelle, research director at the National Center for Scientific Research in Marseille, France. “This result explains the numerous inconsistencies found in the literature.”
Other researchers say this sort of analysis is long overdue.
“This study is to be applauded for highlighting the critical importance of accounting for the effects that different scanners have on the fidelity of brain imaging signals,” says Nicholas Lange, associate professor of psychiatry at Harvard University, who was not involved in the research. “It should give brain scientists pause when making claims based on multi-scanner data.”
Dubious datasets:
Deruelle and her colleagues combed through more than 1,000 images from the Autism Brain Imaging Data Exchange (ABIDE). This database includes MRI data from people with and without autism, compiled from 17 different research centers.
They narrowed their sample to 159 people, about half of whom have autism, from three MRI centers. They restricted the study to right-handed boys and men, aged 8 to 23 years, to avoid differences based on gender and handedness, says Guillaume Auzias, postdoctoral fellow in Deruelle’s lab at Aix-Marseille University in Marseille.
The team used a standard statistical method to parse data on the thickness of the cerebral cortex, the brain’s outer rind, at various locations. This step eliminates any variability from the use of different algorithms to measure the depth. What remains, then, are differences related to scanner quirks or to autism itself.
The finding “should give brain scientists pause when making claims based on multi-scanner data.”
Crunching the numbers to isolate the effects of autism, the researchers found that the brains of people with autism consistently differ from those of their typically developing peers only in the thickness of the motor cortex — a region that controls movement. This finding jibes with multiple studies that link an abnormal thickness in this area with the disorder2.
By contrast, they found that variations among scanners at least partially explain the inconsistencies in reports regarding the thickness of the frontal cortex. Abnormalities in this region, particularly in sections governing decision-making and planning, have been thought to underlie some of the core features of autism.
The evidence so far conflicts on what the abnormality might be. Some studies indicate that the frontal cortex is unusually thick in people with autism, whereas others suggest the reverse. The new analysis shows, however, that the thickness in the frontal cortex does not differ between controls and affected individuals.
The researchers also confirmed that age is a factor. In general, the cerebral cortex thins as the brain matures. One 2014 study reported that in people with autism, the cortex shrinks unusually rapidly in late childhood and then more slowly in adulthood than in controls. But the new analysis indicates no difference in thinning over time between the groups. Again, scanner-related variations can at least partially account for the lack of consensus, Auzias says.
For certain brain regions, such as the insular cortex, which is involved in emotion processing, the researchers found that some scanners register a larger age-related change than others. Depending on the scanner, “the way the cortical thickness evolves with age is not the same,” Auzias says.
The findings indicate that the variations between scanners and the way they are used play an important but complex role in the data they produce.
“We all imagined that this was the case, and ABIDE allows us now to measure the extent of these differences,” says Roberto Toro, neuroscience researcher at the Institut Pasteur in Paris, who was not involved in the research.
Deruelle and her colleagues plan to extend their study to anatomical features such as the depth of cortical folds and the total surface area of the cortex. They expect that some of these results, such as shape of the cortical surface, are likely to be more consistent than cortical thickness. “These surface-related landmarks appear to be much more robust and reliable across MRI centers than the typically used indices such as cortical thickness or volume,” says Deruelle.
Still, any researcher using pooled MRI data needs to control for variability between scanners. One potential strategy would be to test each scanner using a reference substance — say, an inert material of constant volume, geometry and magnetic properties — for calibration purposes, Deruelle says. Ideally, she adds, investigators would also collect anatomical data at various time points from the same people at each site to control for differences.
By joining the discussion, you agree to our privacy policy.